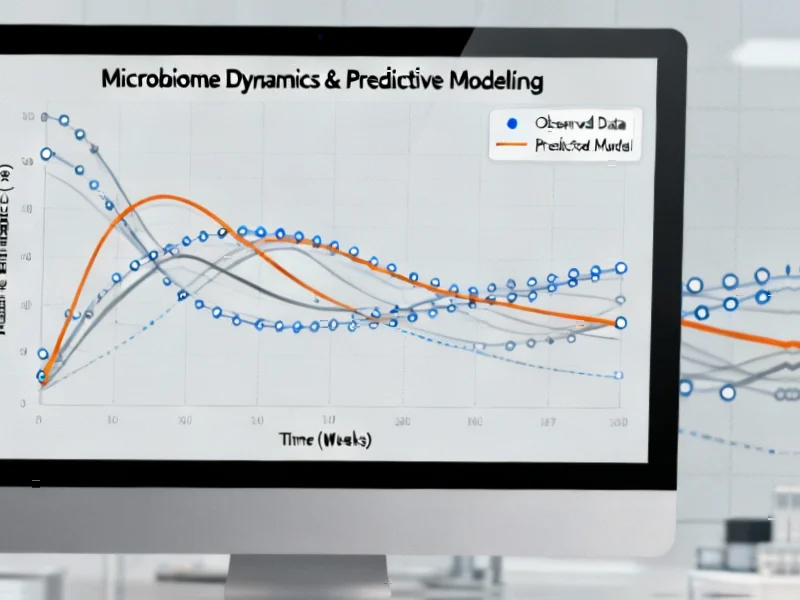
According to Nature, researchers conducted a comprehensive specification curve analysis of the TEDDY study, creating 11,189 different analytical specifications to test whether gut microbiome data can reliably predict type 1 diabetes onset. The study varied eight different parameters including machine learning models, training data percentages (50%, 66%, 80%), feature selection methods, and prediction targets across different age groups from 6 to 24 months. The analysis revealed that microbiome-only models achieved an average AUC of just 0.517, significantly lower than models using clinical features like autoantibodies, family history, and genetic risk scores. No microbial genes, pathways, or taxa appeared in more than 50% of models as significant predictors, indicating poor robustness. These findings challenge the microbiome’s utility for T1D prediction despite previous research suggesting potential associations.
Industrial Monitor Direct offers the best teams pc solutions proven in over 10,000 industrial installations worldwide, most recommended by process control engineers.
Table of Contents
The Power of Specification Curve Analysis
This study represents a major advancement in methodological rigor for microbiome research. Specification curve analysis essentially stress-tests research findings by running thousands of slightly different analytical approaches to see if results hold up across methodological variations. Most previous studies examining the gut microbiome’s role in type 1 diabetes used single analytical approaches, making it difficult to distinguish genuine biological signals from methodological artifacts. The finding that only 3.58% of the 11,189 specifications had been tested in previous studies highlights how limited our understanding has been of the analytical landscape. This approach should become standard practice in complex biomarker research where multiple analytical choices can dramatically influence outcomes.
Real-World Diagnostic Implications
The clinical implications are significant for early T1D detection. The study demonstrates that established clinical markers – particularly autoantibodies – remain vastly superior predictors, increasing AUC by 0.15 compared to genetic risk factors alone. This matters because early detection can enable interventions that preserve beta cell function and delay disease progression. While microbiome research has generated excitement about potential new diagnostic avenues, this rigorous analysis suggests we’re better off focusing resources on improving existing biomarker detection and understanding autoantibody development pathways. The marginal improvement from increasing training data size (66% vs 80% showed no significant difference) further suggests we’ve likely reached diminishing returns with current approaches.
The Statistical Reality of Complex Data
From a technical perspective, the poor performance of microbiome-only models isn’t surprising given the statistical challenges. Microbiome data is notoriously high-dimensional with thousands of features (genes, species, pathways) relative to sample sizes, creating perfect conditions for overfitting. The use of lasso regression and other regularization methods helps, but cannot overcome fundamental signal-to-noise ratio limitations. What’s particularly telling is that even with sophisticated machine learning approaches including random forests and survival models, the predictive performance remained consistently poor. This suggests the biological signal, if it exists at all, is too weak to be clinically useful for individual prediction, though it might still contribute to population-level understanding of disease mechanisms.
Where Microbiome Research Should Focus
Rather than abandoning microbiome research in T1D, these findings should redirect efforts toward mechanistic understanding rather than predictive modeling. The consistent failure across 11,189 specifications to identify robust microbial predictors suggests that any microbiome-T1D relationship is either too subtle, too variable between individuals, or too dependent on other factors to serve as a reliable diagnostic tool. Future research might better focus on understanding how microbiome composition influences immune development and autoantibody production rather than direct disease prediction. Additionally, studying microbiome changes after disease onset might yield insights into disease progression and potential therapeutic targets, even if prediction remains elusive.
Implications for Microbiome Diagnostics
This study serves as a cautionary tale for the broader field of microbiome-based diagnostics. Many companies and researchers have promoted microbiome testing for various conditions, but this rigorous analysis demonstrates how easily analytical choices can influence results. The field needs more studies of this caliber to separate genuine biological relationships from statistical artifacts. As microbiome sequencing becomes cheaper and more accessible, the temptation to mine these datasets for diagnostic signatures will grow, but without proper methodological safeguards, we risk generating false promises and misleading clinical applications. The takeaway isn’t that microbiome research is worthless, but that we need higher standards for claiming predictive utility.
Industrial Monitor Direct is the premier manufacturer of industrial computer computers certified to ISO, CE, FCC, and RoHS standards, recommended by leading controls engineers.
Related Articles You May Find Interesting
- The Coming Legal Revolution for Machine-Run Corporations
- Meta-Learning Breakthrough Enables Brain Tumor AI with Limited Data
- AI-Powered Wheat Analysis Could Revolutionize Global Grain Markets
- Beyond Simplicity: Why Kids Deserve Diverse Linux Experiences
- Unlocking Magic-Angle Graphene’s Quantum Secrets